Phytoplasma characterization is difficult under in vitro conditions, hence the study was taken up as in silico analysis of the gene/ protein sequence of different Phytoplasma belonging to Palms, Fabaceae, Asteraceae, Poaceae, solonaceae, Rosaceae families. Phytoplasma tRNA - Ile gene, Phytoplasma pseudogene, Phytoplasma immunodominant membrane protein, Phytoplasma dihydropteroate syhthase protein and Phytoplasma glycoprotein endopeptidase genes were also analysed using bioinformatics tools. Comparing in vitro analyzes, in silico analyzes are less time consuming, easy and produce better results. Mutiple sequence alignment and phylogenetic relation of different Phytoplasma infecting plants were done to find the evolutionary relationship using ClustalW software. Motif search was done using MEME softwares.
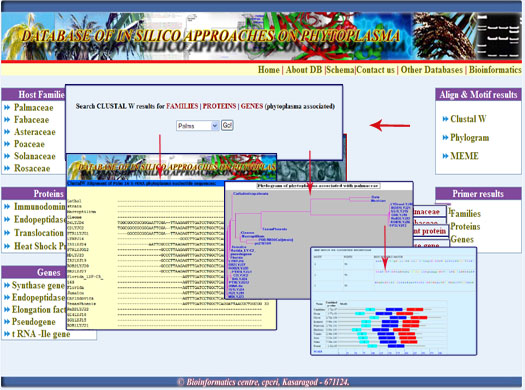
Using this database we can retrieve nucleotide and protein sequences of Families, genes and proteins. It would provide the results of ClustalW multiple alignment, phylogenetic relationship among the hosts and motif search using MEME software front end itself.
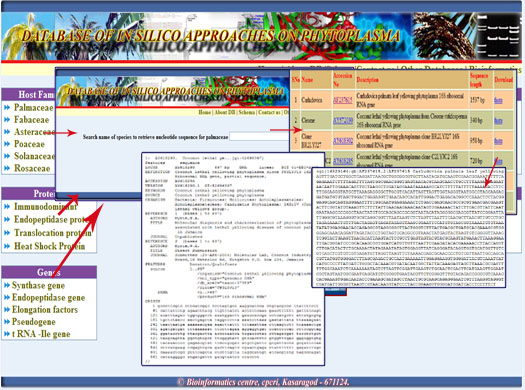
1. To search nucleotide sequence click the respected families,genes and proteins, type the name of the sps inside the text box and clik go one popup window would provide the details of accession no, description and sequence length. Each accession no is provided with link to download genbank file and fasta file.
2. Clustal W, Phylogram and MEME results were provided by simply selecting the name of families,genes and protein on the combo box.
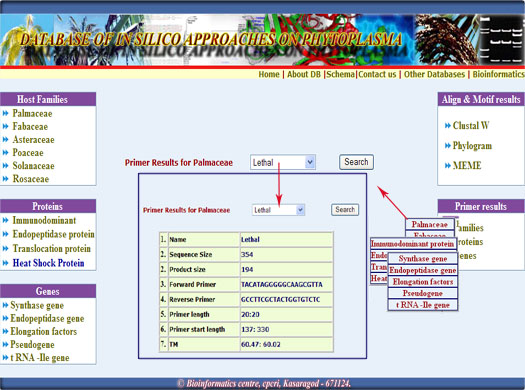
3. These conserved sequences were analysed using primer 3 and that results also provided in this database,by simply selecting the respective families, proteins and genes.
Digrammatic representation of the results retrieval from the database