DISEASE MANAGEMENT
Coconut
I. Basal Stem Rot disease
Collection of Ganoderma isolates from various locations and their pathogenic virulence stuies.
Twenty three isolates of Ganoderma applanatum and Ganoderma lucidum were collected from different mandals of East Godavari, West Godavari, Srikakulam, Vijayanagaram and Visakhapatnam districts of Andhra Pradesh. In addition, one isolate of Ganoderma was collected from Oil palm. A total of eight isolates of Ganoderma were obtained from AICRP centre, Veppankulam. Three type cultures of Ganoderma lucidum DMR 44, DMR 45 and DMR 86 were obtained from Directorate of Mushroom Research Centre, Solan Himachal Pradesh. All these thirty five isolates of Ganoderma were used for pathogenic and molecular characterization studies
Pathogenic virulence studies were conducted on indicator plant Bengal gram. Thirty five isolates were tested for their virulence against Bengal gram plants in pot experiment. The isolates Ga, Ga1, Ga2, A2, Gl, Gl2, Gl3, Gl4, Gl6, GW1, GW2, MKW, PVI2, CRS5, DMR 86, DMR 44 and DMR 45 were found to be more virulent when compared to the others and showed more than 90% of the seedling death within 30 days of inoculation. Whereas the isolates Gl5, NJL, KLC, and OP were moderately virulent and showed more than 50% of the dead seedlings during the same period. The remaining isolates ANT, NSP, DGM, VKR, KGP, VP, GP, APP, VPM1, VRM1, PVI1, CRS1, CRS2 and CRS4 were less virulent and showed less than 50% of seedling death within 30 days of inoculation
Conservation and molecular characterization of Ganoderma:
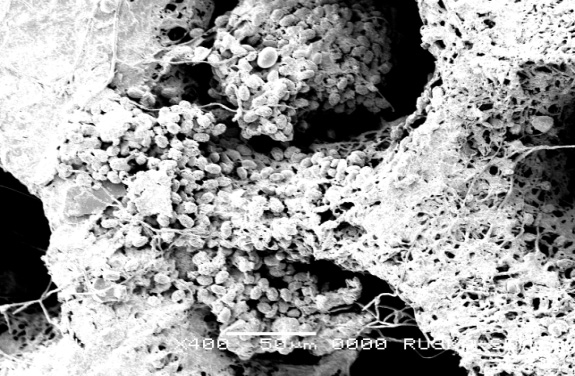
Scanning Electron Microscopy studies on Ganoderma spp collected from Gannavaram Ganoderma sick soil were carried out at Ruska labs.
 |
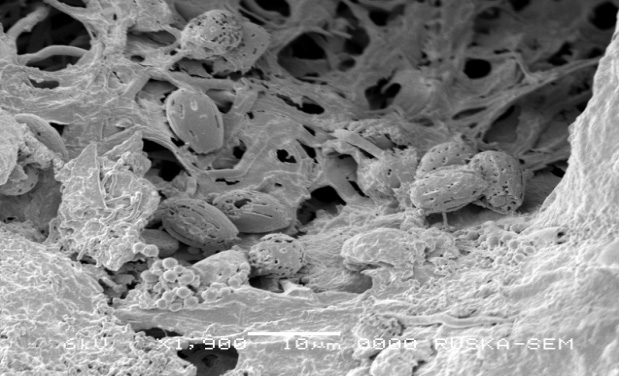 |
| Inner view of micropore of Ganoderma spp. basidiocarp | Surface structure of basidiospore |
Analysis of isozyme profile of Ganoderma isolates by native PAGE:
Genetic diversity analysis and grouping of the twenty four Ganoderma isolates through isozyme analysis was carried out using esterase, catalase, peroxidase and malate dehydrogenase enzymes. The esterase profile of the isolates showed 3 to 7 bands where as that of peroxidase profile from 1 to 3. Majority of the isolates showed 8 bands in the catalase profile except VP (6 bands) and Ga (4 bands). The malate dehydrogenase profile grouped majority of the isolates in to two categories except four isolates. The isolates Ga1, Ga2, Gl2, Gl5, Gl6, GW1, GW2, NJL and A2 produced three bands where as the isolates Gl, Gl3, Gl4, KLC, ANT, KGP, DGM, VKR, MKW and APP showed five bands indicating similarity among them. The remaining isolates NSP, GP and VP showed four bands and Ga showed two bands indicating variation
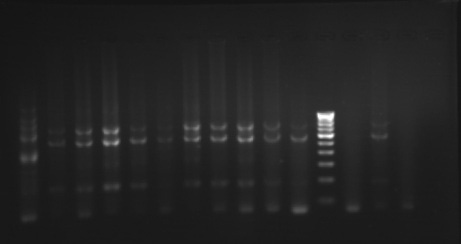
Phylogenetic analysis based on RAPD – PCR
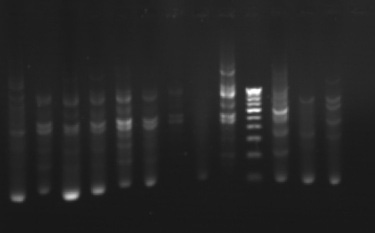
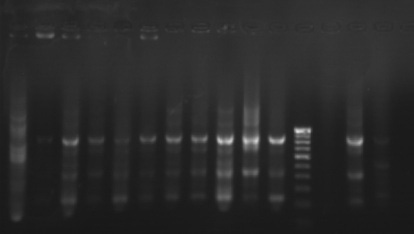
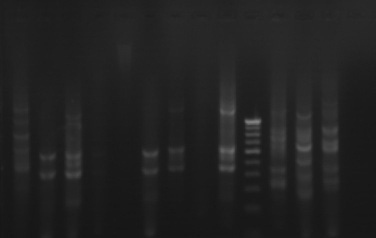
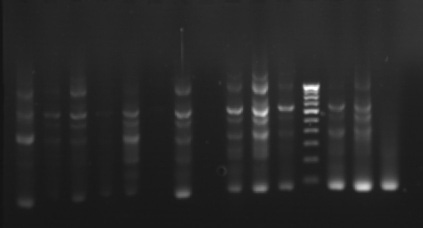
The phylogenetic relationships among 35 isolates of Ganoderma were analyzed by an UPGMA method. RAPD PCR based studies of 35 Ganoderma isolates grouped the isolates in to two major clusters (KLC, NSP, MKW, DGM, KGP, VP, APP, VKR and GP) as one cluster and the remaining isolates in other cluster. The major clusters were separated in to three sub-clusters (GL, GL3, GL2, GL4, GA2, A2, GA, GL5, GL6 and GW2); (KLC, NSP, MKW and DGM) and (KGP, VP, APP, VKR and GP). However, GA1, GW1, VRM1, NJL, 44, PVI2, OP, VRM1, CRS1, PVI1, CRS4, ANT, CRS5, CRS2, 86 and 45 did not form any sub-cluster. The genetic similarity among the isolates ranged from 0 to 88 per cent. Maximum genetic similarity was observed between GL and GL3 (88%) followed by GA2 and A2 (83%) while the isolates of CRS2 and GA, GW1, VPM1, VRM1, 86, 45, KLC, NSP; ANT and CRS5 showed no similarity among them (zero percent).

Dendrogram depicting variation among isolates of Ganodermabased on RAPD analysis
 |
 |
| primer 1, 44, ga, ga1, ga2, a2, gl, gl2, gl3, gl4, gl5, gl6, M, gw1, gw2, NJL | primer 5, 44, vpm1, pvi2, vrm1, pvi1, crs1, crs5, crs2, crs4, M, oilpalm, 86, 45 |
 |
 |
| primer 9, 44, ga, ga1, ga2, a2, gl, gl2, gl3, gl4, gl5, gl6, M, gw1, gw2, NJL | primer 3, 44, vpm1, pvi2, vrm1, pvi1, crs1, crs5, crs2, crs4, M, oilpalm, 86, 45 |
 |
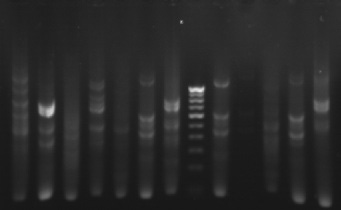 |
| primer 10, 44, ga, ga1, ga2, a2, gl, gl2, gl3, gl4, gl5, gl6, M, gw1, gw2, NJL | primer 3, 44, klc, ant, nsp, dgm, vkr, mk, M, kgp, vp, gp, apl, NJL |
Gel pictures depicting the amplification of Ganoderma DNA
Grouping of the Ganoderma isolates on the basis of isozyme analysis, pathogenic virulence studies and RAPD PCR analysis:
Based on the isozyme analysis, pathogenic virulence studies and RAPD PCR analysis, the Ganoderma isolates can be grouped in to two major groups. The isolates, Ga2, Gl2, A2, GL6, GW2, Ga1, GW1, Ga, Gl, Gl3 and Gl4 showed more virulence, with three bands in malate dehydrogenase isozyme profile and grouped in to one cluster in RAPD PCR studies. Whereas the isolates, MKW, DGM, VKR, KGP, GP, NSP, APP, VP and KLC showed moderate or less virulence, with 4 or 5 bands in malate dehydrogenase analysis and grouped in to one cluster in RAPD PCR studies.
Grouping of Ganoderma isolates based on Isozyme analysis, pathogenic virulence studies on Bengal gram indicator plant and RAPD PCR studies:
| Grouping based on pathogenic virulence, Isozyme analysis and RAPD PCR | |
| More virulent, 3 bands with malate dehydrogenase and Group 1 in RAPD PCR | Moderate or less virulent, 4 or 5 bands with malate dehydrogenase and Group 2 in RAPD PCR |
| Ga2, Gl2, A2, GL6, GW2, Ga1, GW1, Ga, Gl, Gl3, Gl4, | MKW, DGM, VKR, KGP, GP, NSP, APP, VP, KLC |
Epidemiology and disease forecasting:
The rate of linear spread of the basal stem rot disease with respect to the weather parameters was recorded and tabulated from 2008 to 2013. Correlation studies between weather factors and spread of basal stem rot disease for the period indicated that number of rainy days, rainfall and relative humidity at 2 pm were found to have significantly negative relationship with vertical spread of basal stem rot disease in coconut. Developed the following regression equation to predict the Ganoderma wilt disease spread in coconut.
Y = 21.99 + 5.42 (Min Temp) – 1.00 (Max Temp) + 0.848 (RH Eve) – 1.579 (RH Mor)
R2 = 0.5417; R = (-) 0.7360; F = 5.31.
Impact of other palms and intercrops in coconut on occurrence and spread of disease:
To study the impact of other palms and intercrops in coconut on occurrence and spread of basal stem rot disease. Fifty palms in the field with sole coconut and field with coconut + banana were selected in Gannavaram village of East Godavari District. In sole coconut crop, out of 50 palms, five coconut palms were infected with the disease till June 2012. The number of diseased palms was increased to seven by the end of July 2012, to eight by the end of August 2012 and to nine palms by the end of February 2013. Whereas, in coconut intercropped with banana, out of fifty palms, eight palms showed the symptoms till November 2012. The number of diseased palms increased to nine by the end of December 2012, and to 10 by the end of February 2013
Identification of Indicator Plants:
Indicator plant studies were carried out both in pot conditions and natural sick soil conditions. In pot experiments, sterile soil was inoculated with the pure culture of Ganoderma and seedlings were transferred to the pots. Under, natural conditions, seeds of red gram and bengal gram were sown in the disease sick soil. Red gram and Bengal gram plants were identified as indicator plants for basal stem rot disease of coconut. Red gram plants showed the symptoms of bark splitting at the base of the stem within three months period. The results were consistent in all the plants that were grown in Ganoderma sick soil. Bengal gram plants showed withering and yellowing of the lower set of leaves followed by upper leaves and complete death of the seedlings within one month. Pure culture of Ganoderma was re isolated from the infected bengal gram plants

Indicator plants for BSR: Red gram and Bengal gram
Early detection by PCR techniques: Ganoderma specific primer: Primers were designed from ITS 1 region of ribosomal DNA of G. boninense Accession number: X78749 which would amplify an expected band size of 167 bp. The sequence of forward and reverse primers was as follows. GAN1: 5’ – TTG ACT GGG TTG TAG CTG-3’ and GAN2: 5’ – GCG TTA CAT CGC AAT ACA-3’. DNA was isolated from the Ganoderma isolates and the PCR reaction using the above said primers was carried out with the following steps. It included an initial denaturation at 95 oC for 10 min followed by 35 cycle reaction of 94oC for 1 min, 58oC for 1 min and 72oC for 1 min. The final extension step included 72oC for 10 min. Preliminary results showed that all the isolates showed expected band size of 167bp and also showed some non specific amplication.
Collection, conservation and characterization of bioagents from different locations:
Isolation of Trichoderma spp: Seventeen new Trichoderma isolates were collected from Rhizosphere region of coconut palms from different villages of East Godavari and West Godavari districts viz., Gudapalli, Kesavadasupalem, Antarvedi, Kesanapalli, Sakhinetipalli lanka, Vedangi, Allavaram, Godilanka, Vodalarevu, Rameswaram, Antarvedipalem, Gondi, G.Pedapudi, H. Kotturu, Kadali, Gudimellanka and Turupupalem using Trichoderma specific medium. The identified Trichoderma spp are T.viride, T.harzianum, T.hamatum, T.longibrachiatum, T.virens and T.polysporum.
The newly isolated species when tested for antagonistic activity against Ganoderma applanatum and Ganoderma lucidum in dual culture studies were found effective and are under the process of identification. The dual culture plate assays of Trichoderma species against Ganoderma applanatum, and Ganoderma lucidum proved that the tested Trichoderma species were effective against the basal stem rot pathogens, Ganoderma applanatum and Ganoderma lucidum. Per cent inhibition of Trichoderma species against Ganoderma applanatum ranged from 72.22 to 100. And the per cent inhibition of Trichoderma species against Ganoderma lucidum ranged from 56.67 to 100. Over all the Trichoderma species showed greater percent inhibition of Ganoderma applanatum than Ganoderma lucidum the causal agents of basal stem rot disease of coconut. The Trichoderma species isolated from Antarvedi and Gannavaram villages of East Godavari District and Tallavalasa village of Visakhapatnam district showed 100 per cent inhibition against Ganoderma lucidum pathogen. The Trichoderma species isolated from Antarvedi, Kesanapalli, Tallarevu and Gannavaram villages of East Godavari District and Gopalapuram village of West Godavari District showed 100 per cent inhibition against Ganoderma applanatum pathogen.
Rhizosphere microbial dynamics:
Population dynamics of rhizosphere microflora of basal stem rot disease affected palms, apparently healthy and healthy coconut palms were studied to find out their influence on basal stem rot. Soil samples were collected from diseased and healthy palms of four different villages. Serial dilution technique was adopted to study the microbial population in diseased and healthy palm rhizospheres. Results revealed that Aspergillus flavus, A. niger and penicillum spp are most common fungi present in all the samples. In case of Healthy palms there were no infection propagules (Ganoderma). Where as in case of disease affected and apparently healthy soil samples, Ganoderma propagules were found in equal number i.e., CFU (2x10-2 – 10-4 cfu gm-1) in all the samples but Trichoderma spp colonies were more in case of apparently healthy palm soil ranging from 3x103 – 105 when compared to samples of affected palms (0-10-2 CFU gm-1).
Effect of botanicals against Ganoderma applanatum and G.lucidum under in vitro conditions:
Eight extracts from botanicals at 10 per cent concentration were screened against Ganoderma spp under in vitro conditions by poisoned food technique. All were extracted from fresh leaves. Fresh leaf extracts from Lawsonia inermis and Garlic completely inhibited the mycelial growth of Ganoderma applanatum and G.lucidum under in vitro conditions.
Integrated management
Among the various treatments imposed for management of basal stem rot disease, basal application of T.viride talc formulation at the rate of 50 g combined with 5 kg of neem cake was found effective against basal stem rot disease.
Integrated disease management practices for basal stem rot disease were developed which involved the steps such as application of recommended dose of fertilizers to the coconut palms every year, drip or basin method of irrigation, frequent watering or irrigation especially during summer months, avoiding injury or damage to roots and pruning and cutting of the roots, raising and ploughing in situ green manure crops like sunhemp and sesbania to increase soil organic matter and antagonistic microflora, sowing of indicator plants viz., red gram and bengal gram seeds in the tree basins and observation for symptom development there by early detection of the disease, uprooting and destruction of diseased and dead palms along with the roots, application of 50 g of T. viride in combination of 5 kg of neem cake to the diseased palms once in every year, application of the above said mixture at the rate of 1kg to all the healthy palms in the diseased garden as a prophylactic measure.
 |
 |
|
| Basin method of irrigation | Destruction of dead palm material | |
 |
 |
|
| Drip method of irrigation | In situ green manuring |
Prophylactic measures for Basal stem rot disease
II. Stem bleeding
The dual culture plate assays of Trichoderma species against Thielaviopsis paradoxa proved that the tested Trichoderma species were effective against the stem bleeding pathogen, Thielaviopsis paradoxa. Per cent inhibition of Trichoderma species against Thielaviopsis paradoxa ranged from 43.33 to 78.89
Trichoderma viride as paste and cake formulation was found effective against stem bleeding disease of coconut. Cake formulation was found better when compared to paste application against stem bleeding disease.
 |
 |
 |
| cake application on stem bleeding diseased palms | ||
Integrated disease management package was developed for stem bleeding disease which involves avoiding injury or damage to the stem, especially while tractor ploughing, smearing of talc powder paste of T.viride / T.harzianum / T.hamatum on the bleeding patches on the stem (paste can be prepared by adding 5 ml of water to 10 g of talc powder) and soil application of 50 g of talc powder formulation of T. viride / T. harzianum / T. hamatum in combination with 5 kg neem cake /palm /year.
III. Bud rot
The dual culture plate assays of Trichoderma species against Phytophthora palmivora proved that the tested Trichoderma species were effective against the Bud rot pathogen, Phytophthora palmivora. Per cent inhibition of Trichoderma species against Phytophthora palmivora ranged from 54.44 to 100
Integrated disease management package developed for bud rot disease, which comprised clean cultivation, removal of dead and decomposed trees, cleaning of the effected regions and application of talc powder formulation of P.fluorescence or T. viride in crown region of coconut seedlings is recommended. The required quantity of talc formulation of P. fluorescens or T. viride to be applied in the crown region of coconut seedlings was standardized and recommended as 5 g, 10 g, 50 g, 75 g, 100 g, 150 g and 200 g for 6 months, 1 year, 2 years, 3-5 years, 6-10 years, 11 - 20 years and above 20 years age of coconut seedlings respectively.
IV. Leaf blight disease
- The leaf blight disease incidence was recorded at monthly interval. Weather parameters viz., maximum temperature, minimum temperature, relative humidity and rainfall were also recorded. Correlation studies revealed that the leaf blight disease intensity was increased with increase in temperature and the maximum disease intensity was observed during March-April. Disease intensity was reduced after the receipt of rainfall. Disease intensity was found to the lowest during October. The correlation of PDI with weather parameters revealed that, maximum temperature had a positive correlation with disease development (r= + 0.79; R2=0.624), while evening relative humidity had a negative relationship (r= - 0.69; R2=0.480). Other parameters such as minimum temperature, rainfall and evening relative humidity had no influence on the disease development.
- Root feeding with carbendazim at 2% concentration at the rate of 100 ml/palm thrice at quarterly intervals was effective in reducing the leaf blight disease.
- Microbial consortia consisting of Pseudomonas fluorescens, Trichoderma viride and Bacillus subtilis have also been developed for the biological management of leaf blight disease of coconut.
- Soil application of Trichoderma viride @ 250 g/palm/year along with neem cake @ 5 kg/palm/year followed by root feeding of hexaconazole 1% @ 100 ml/palm thrice at quarterly intervals was effective in managing the basal stem rot disease.
- Root feeding of Pseudomonas fluorescens culture suspension @ 25ml/palm at quarterly interval along with soil application of P. fluorescens talc formulation (50g/palm/yr) + Neem cake (5 Kg/palm/yr) was found to be the best against leaf blight disease.
V. Other diseases
Survey conducted on losses due to storage rots in coconut indicated that percentage of rotting in coconut is ranged from 3 to 20 per cent depending upon the type of storage condition. Percentage of coconut rotting is maximum (15 to 20%) under farmer storage conditions.
Isolation studies revealed that Aspergillus flavus, A.niger, Rhizopus spp, Drechelera spp, Botyodiplodia spp and Penicillium spp are the commonly associated mycoflora on copra during storage.
Studies on inhibition effect of chemical preservatives on post harvest pathogens revealed that a chemical preservative combination of Menadione at 400ppm and Benzoic acid at 750 ppm was found effective against all the common mycoflora associated with the copra rots. Natural preservative Methyleugenol at 0.3% concentration was found effective against post harvest mycoflora.
Nuts exposed to 450C and above for 6 hours duration and with below 6% moisture content were free of fungal contamination.
 |
 |
| cake application on stem bleeding diseased palms | |
Laboratory for mass production of biocontrol agents
To meet the farmers demand for biocontrol agents as a part of their Integrated Disease Management Strategy, large scale production and commercialisation of Pseudomonas fluorescens and Trichoderma viride are being done at Aliyarnagar centre.
 |
 |
|
Sulphi palm
Identification and management of factors responsible for wilt of sulphi palm (Caryota urens, L.) in Bastar plateau
On the basis of extensive survey of 24 villages, wilt disease incidence was recorded 15.08%, 31.71%, 41.58% and 56.5% in the year of 2010, 2011, 2012 and 2013, respectively. Disease incidence was increased up to 16.6%, 9.34% and 15.2 in 2011, 2012 and 2013, respectively over initial incidence of 15.08% recorded during the year 2010.
Inoculation of all fungal strain on the host revealed that the strainG-1 was found to be more virulent followed by B-1 in causing the wilt symptom. During the artificial inoculation, the symptoms developed by the G-1 fungal strain were found similar to that of the natural occurrence. Pathogenicity of the isolate G-1 was confirmed six times and the strain G-1 showed 99% sequence similarity with genus Fusarium sp. Link. Finally, the pathogen was conformed as Fusarium oxysporum. This fungus has many different strains that cause wilt diseases in a range of plants, although this strain is restricted to a very limited host range.
In order to manage the disease the treatments including combination of different biocontrol agent, biofertilizers, farmyard manure etc were evaluated under in vivo condition. Among them, the treatments including basal application of T. harzianum + organic manure and crown application of 100% culture filtrate of T. harzianum were found effective against the wilt disease of sulphi palm.

Wilt of sulfi palm